
Introduction
Hugging Face is currently the most popular AI community globally, fostering innovation and collaboration among data scientists and businesses in various aspects such as models, datasets, and applications. Hugging Face hosts a diverse range of datasets that serve not only as samples for learning and practice but also as foundational sources for enterprise model data.
In order to maximize the utility of these datasets, it is often necessary to cleanse the raw data within them and archive it in a data lake to provide a unified access point. Databend is designed with ETL/ELT-centric workflows in mind, allowing for immediate cleansing, transformation, and merging of data upon loading. It supports a wealth of structured and semi-structured data types, enabling direct querying and analysis of raw data in multiple formats. The enterprise edition further supports advanced features such as virtual columns and computed columns, ensuring that the data is always ready for use.
In this article, we will demonstrate how to effortlessly access datasets hosted on Hugging Face using Databend and perform simple yet efficient analysis and processing with SQL. Additionally, the examples in this post include a white-box model implemented in SQL, showcasing how to conduct category predictions within the data warehouse and validate the model's accuracy.
Accessing Hugging Face Datasets
Hugging Face offers a publicly accessible REST API, which supports the automatic conversion of dataset files to Parquet upon upload, enabling easy integration with databases and data analysis tools.
The dataset page is presented below, where through the Dataset card, one can gain an intuitive understanding of the data and obtain a preview, while the Files and Versions page helps to understand the directory structure and revision history.
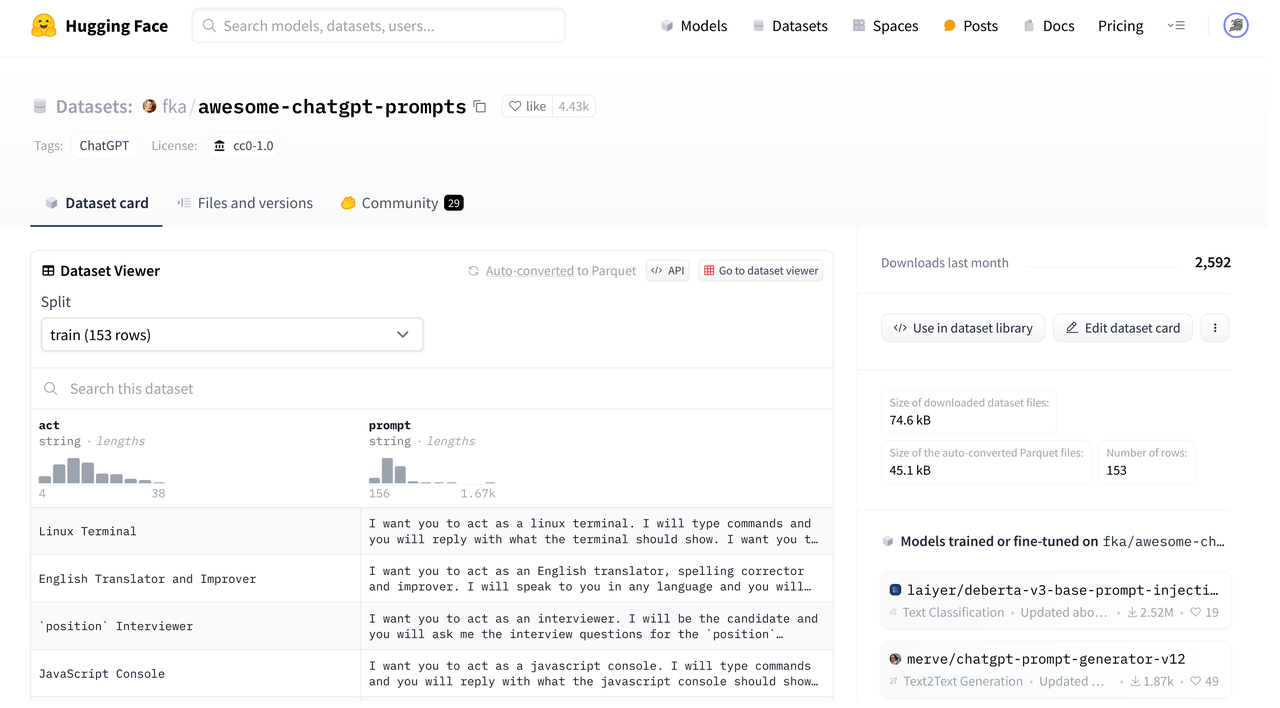
Databend provides several ways to integrate with Hugging Face, and in the following sections, we will use
fka/awesome-chatgpt-prompts
External Stage/Location
Many databases now support direct access to remote data files via a remote URL. This mode is applicable to Hugging Face, as well as other file services that enable HTTPS access. However, direct access via remote URLs has some drawbacks, such as unexpected responses, incorrect escaping and encoding/decoding, and the need for complex JSON parsing to locate files.
In Databend, you can directly query the datasets in the Hugging Face or mount the filesystem as an external Location or Stage, effectively avoiding access issues and facilitating data querying.
For Hugging Face, the applicable externalLocation parameter is as follows:
externalLocation ::=
"hf://<repo_id>[<path>]"
CONNECTION = (
<connection_parameters>
)
URI Format:
hf://{repo_id}/path/to/file
repo_id
fka/awesome-chatgpt-prompts
- : The type of Hugging Face repository, with the default being dataset. Available options are dataset, model.
repo_type - : The revision version in Hugging Face, defaulting to main. It can be a branch, tag, or commit in the repository.
revision - : The API token for Hugging Face.
token
Querying the raw CSV data file as an external Location
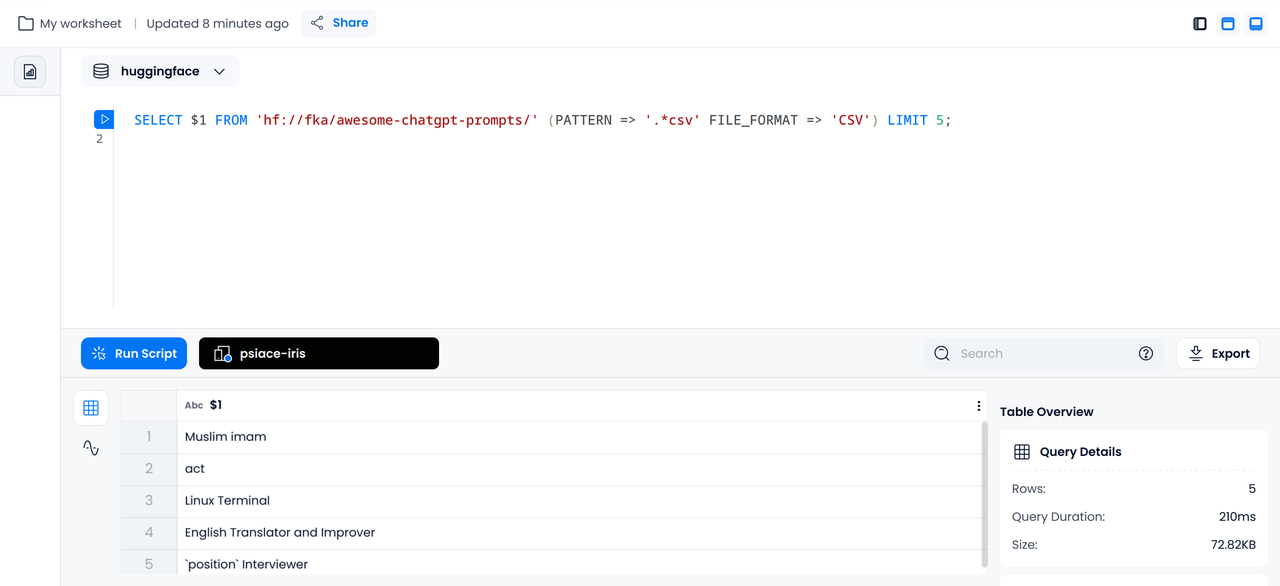
The following example shows how to use Databend to directly query the raw CSV data file of
fka/awesome-chatgpt-prompts
SELECT $1 FROM 'hf://fka/awesome-chatgpt-prompts/' (PATTERN => '.*csv' FILE_FORMAT => 'CSV') LIMIT 5;
Querying the transformed Parquet data file as an external Stage
As mentioned earlier, Hugging Face provides Parquet files that have been converted, and Databend also supports mounting the Hugging Face filesystem as an external Stage. The following example demonstrates its usage:
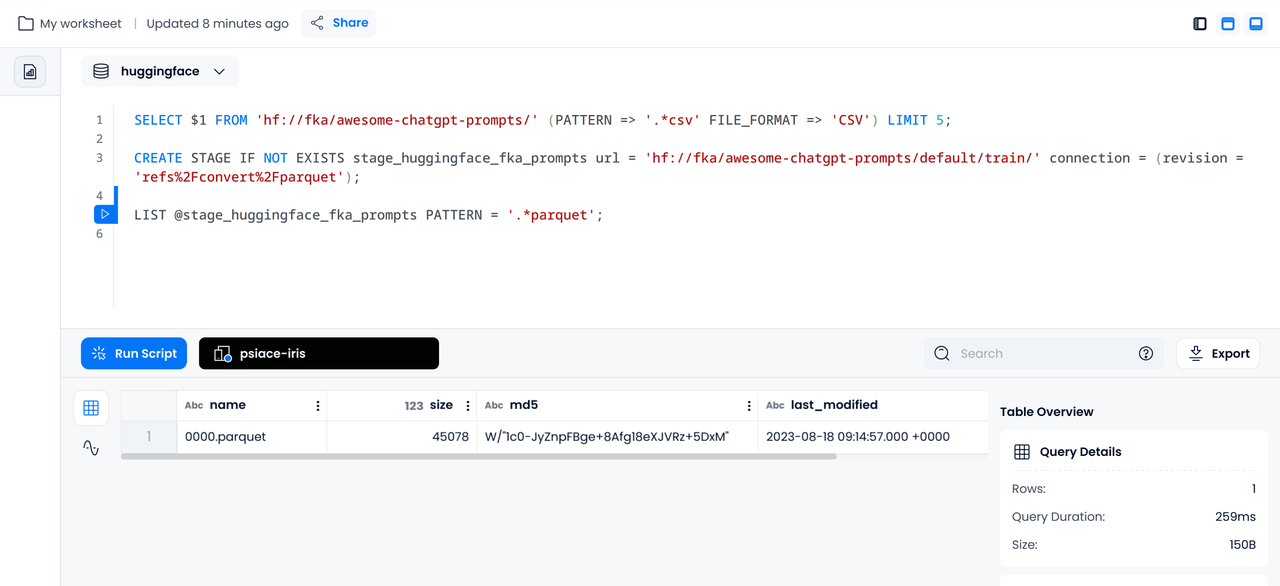
Creating a Stage
For this example, the converted files are located in the
/default/train/
refs%2Fconvert%2Fparquet
CREATE STAGE IF NOT EXISTS stage_huggingface_fka_prompts url = 'hf://fka/awesome-chatgpt-prompts/default/train/' connection = (revision = 'refs%2Fconvert%2Fparquet');
Listing Corresponding Files
We can use
PATTERN
LIST @stage_huggingface_fka_prompts PATTERN = '.*parquet';
Querying Data
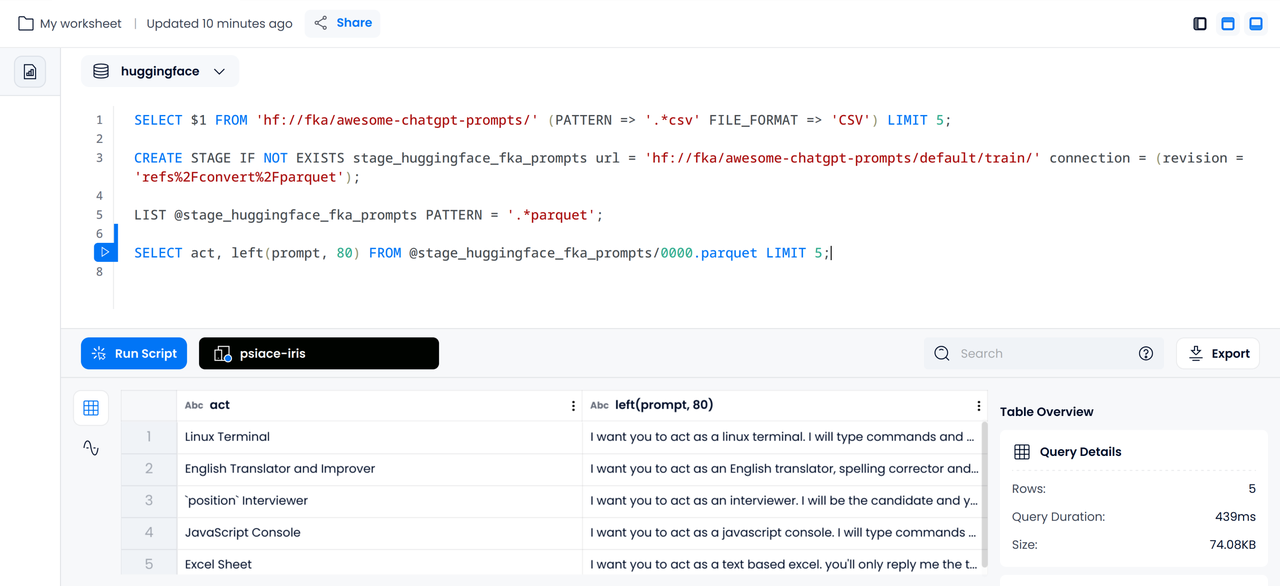
Since we have already obtained the file's information from the CSV file, when querying the Parquet file, we can directly select its columns. Considering that the prompt is inconvenient to display in full in the terminal, here it is truncated to 80 characters.
SELECT act, left(prompt, 80) FROM @stage_huggingface_fka_prompts/0000.parquet LIMIT 5;
Data Science with SQL
In the previous section, we discussed how to perform simple queries on remote datasets at Hugging Face. In this section, we will look at a specific example, exploring data science using SQL.
For ease of understanding, we select the commonly used iris classification dataset for beginners in data science as an example. The Iris dataset appears in R.A. Fisher's classic 1936 paper The Use of Multiple Measurements in Taxonomic Problems and can also be found in the UCI Machine Learning Repository, making it very suitable for demonstrating simple classification models.
The Hugging Face dataset used in this article is located at https://huggingface.co/datasets/scikit-learn/iris .
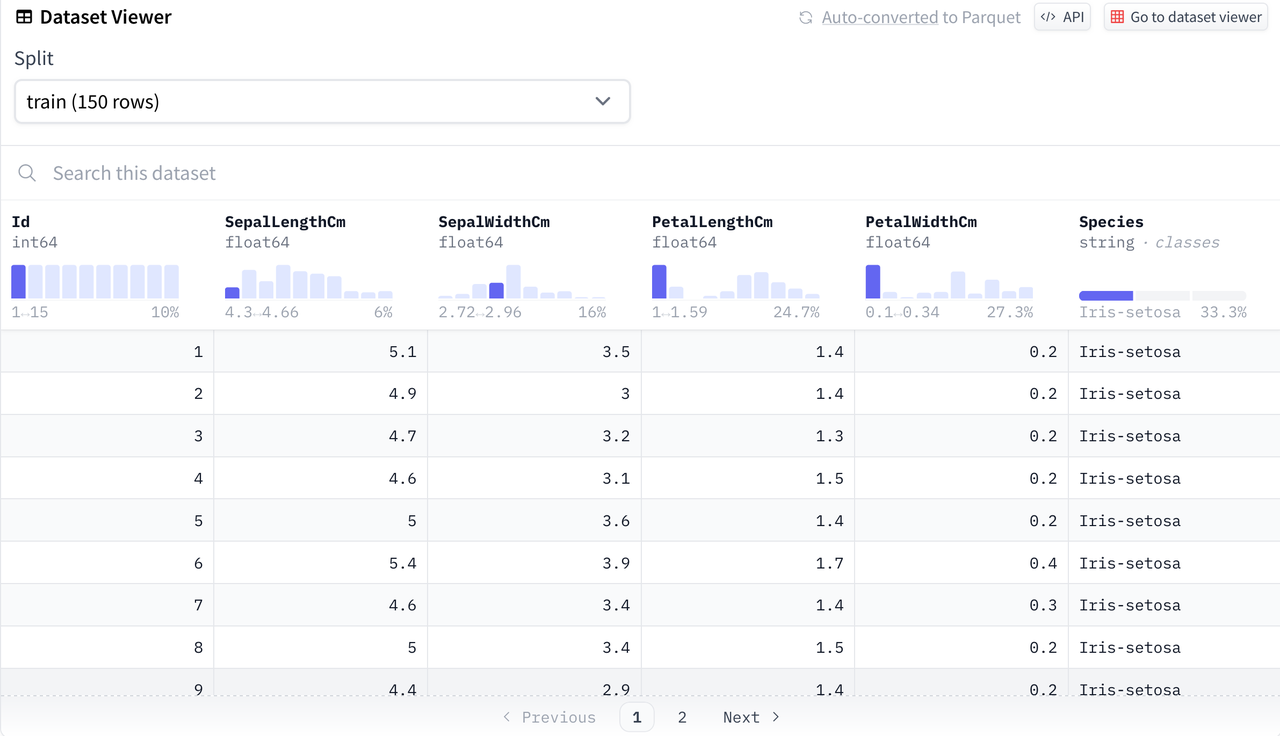
Mounting Data
We will mount the dataset as an external Stage in Databend:
CREATE STAGE IF NOT EXISTS stage_iris url = 'hf://scikit-learn/iris/default/train/' connection = (revision = 'refs%2Fconvert%2Fparquet');
LIST @stage_iris PATTERN = '.*parquet';
SELECT * FROM @stage_iris/0000.parquet LIMIT 5;
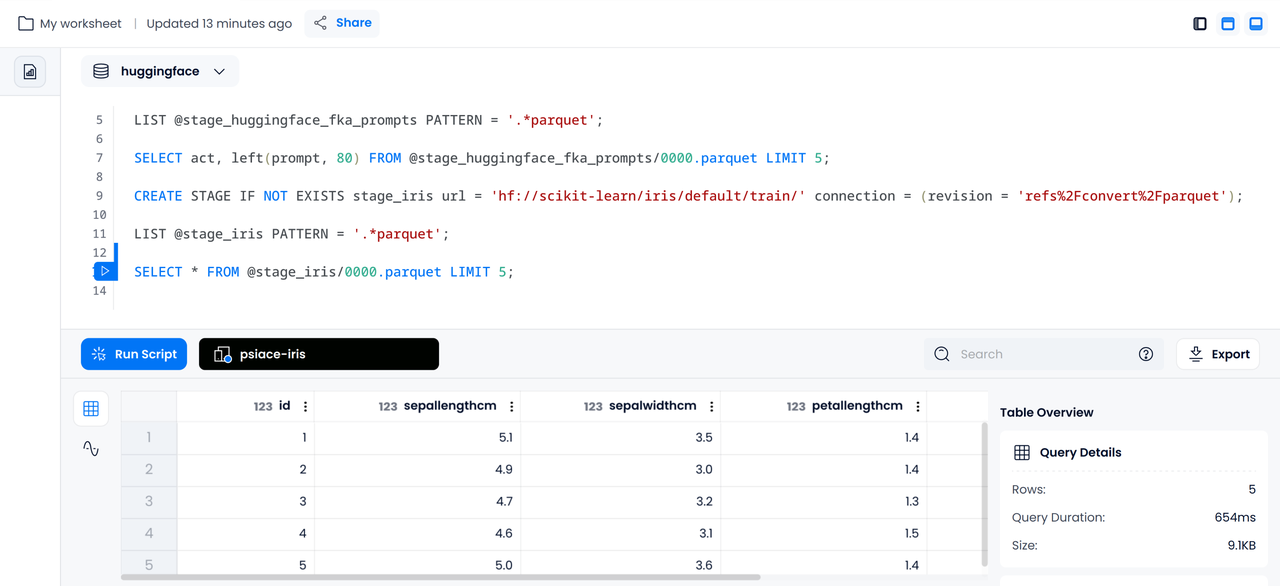
Data Cleaning
The data in the external Stage is equivalent to the raw data. To meet the standards and needs for use, you can clean the data during import. For the Iris dataset, typical cleaning operations include:
- Removing the column.
id - Converting the column to categorical labels, i.e., integer form.
species - Converting feature columns from to
float64.float32 - Renaming feature columns to snake case.
In Databend, data cleaning can be done during loading using the
COPY INTO
Creating the Corresponding Table
This statement creates a table in Databend that aligns with the dataset.
CREATE TABLE iris (
sepal_length FLOAT,
sepal_width FLOAT,
petal_length FLOAT,
petal_width FLOAT,
species INT
);
Using COPY INTO to Import Data
The following statement cleans and imports the data in a single command:
COPY INTO iris
FROM (
SELECT
to_float32(t.sepallengthcm),
to_float32(t.sepalwidthcm),
to_float32(t.petallengthcm),
to_float32(t.petalwidthcm),
CASE
WHEN species = 'Iris-setosa' THEN 1
WHEN species = 'Iris-versicolor' THEN 2
WHEN species = 'Iris-virginica' THEN 3
ELSE NULL
END AS species
FROM @stage_iris t
)
FILE_FORMAT = (TYPE = PARQUET)
PATTERN = '.*parquet';
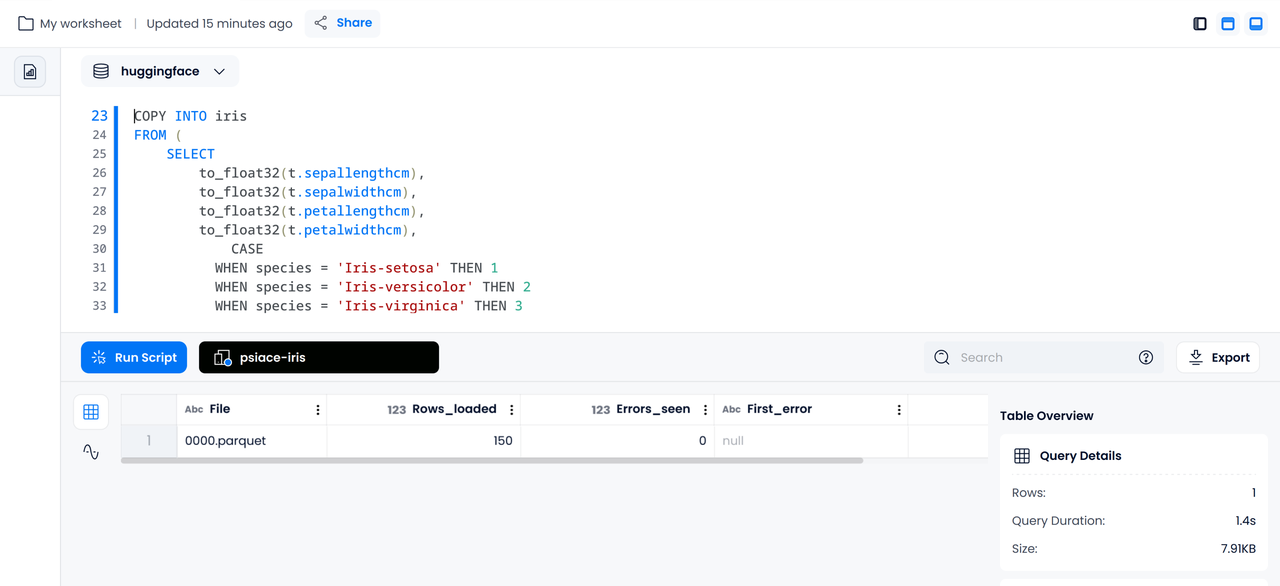
Basic Statistics
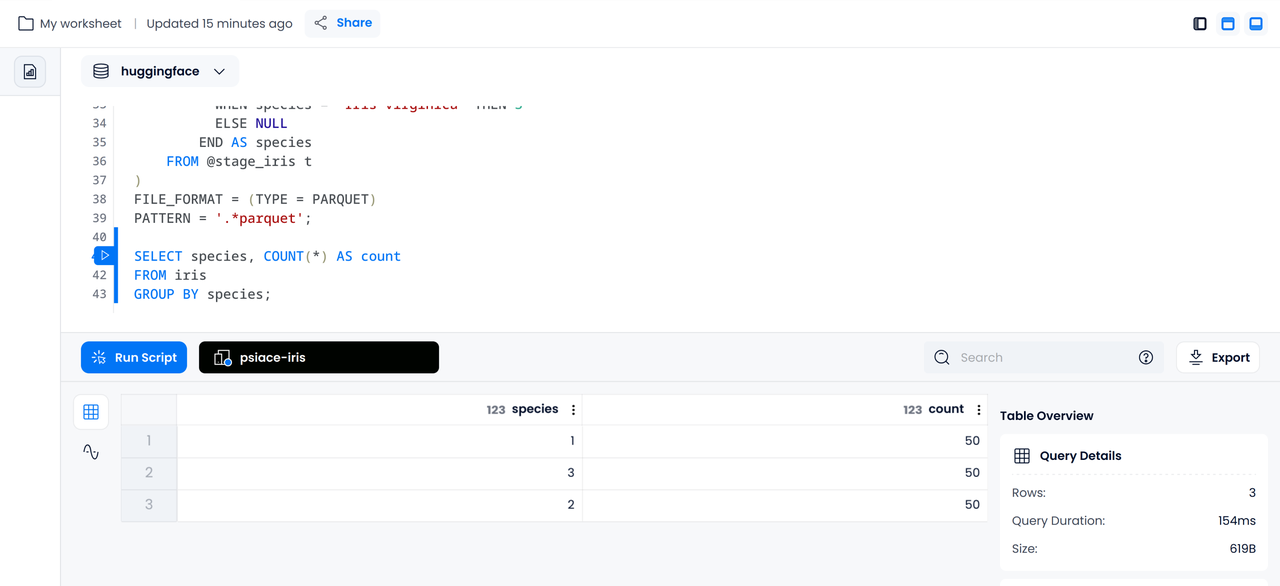
The following statement helps us figure out how many items in each specie based on the cleaned data:
SELECT species, COUNT(*) AS count
FROM iris
GROUP BY species;
Basic Analysis
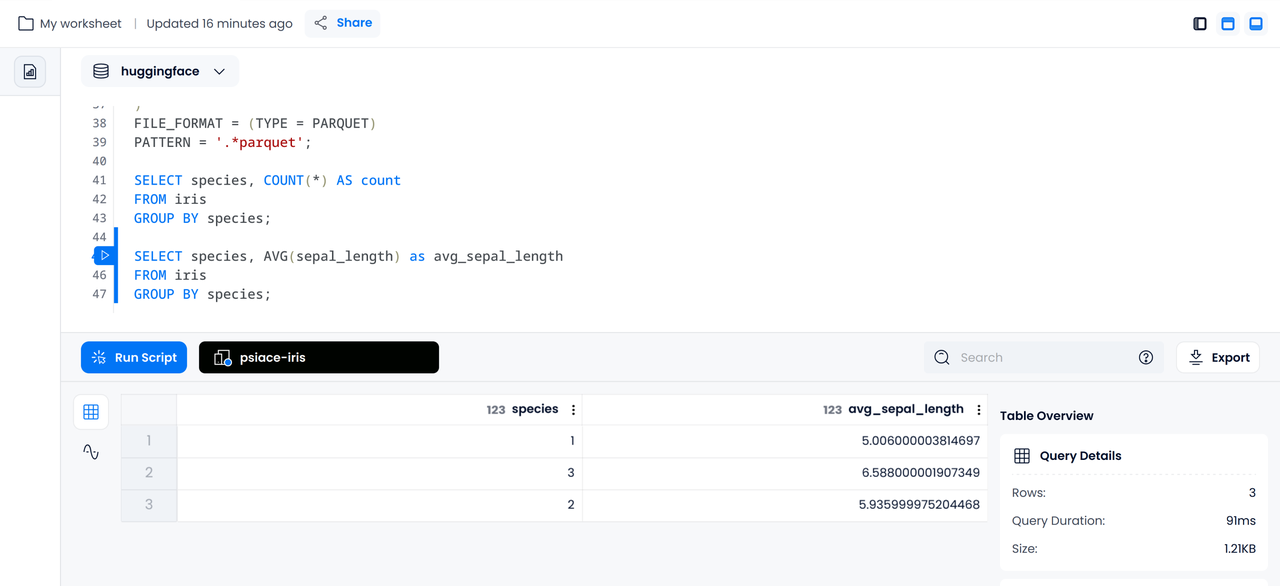
By executing the following statement, we can get the average sepal length for each type of iris:
SELECT species, AVG(sepal_length) as avg_sepal_length
FROM iris
GROUP BY species;
Using SQL to Predict Iris Categories
Mocking up a simple white-box model with SQL can intuitively feel the prediction standards of the model and avoids interacting with model services.
The example below is for demonstration purposes only and does not comply with the rigorous training, validation, and application data science process.
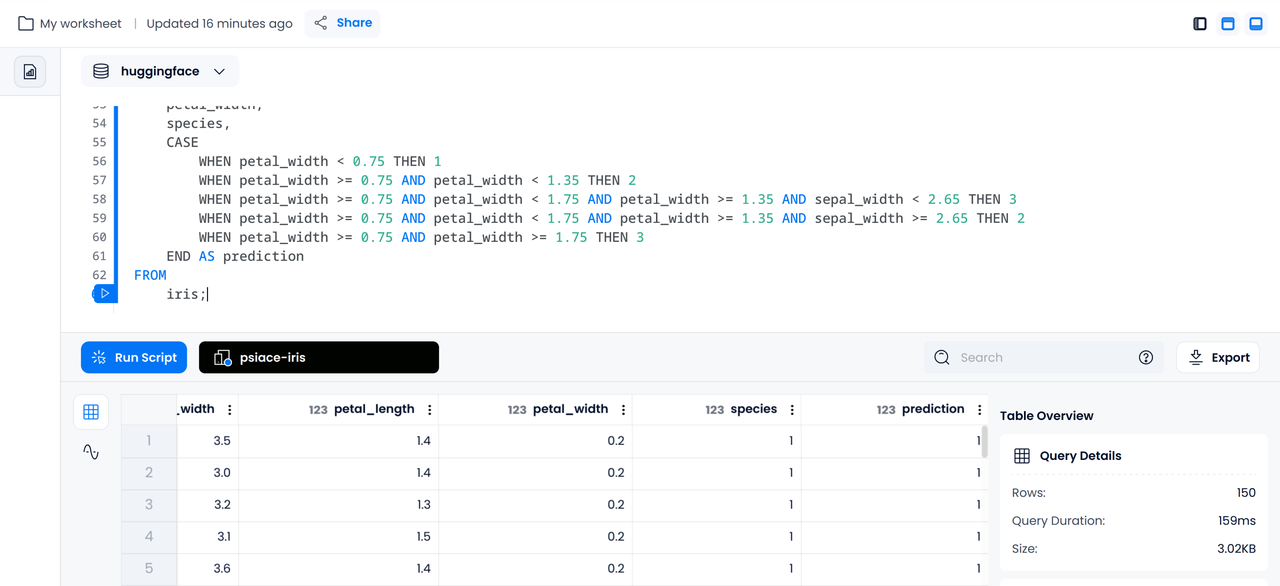
Let's assume we have received criteria for categorizing irises by their features from the work of some data scientists. Now, let's "create a model in Databend" using the following CASE WHEN statement:
SELECT
sepal_length,
sepal_width,
petal_length,
petal_width,
species,
CASE
WHEN petal_width < 0.75 THEN 1
WHEN petal_width >= 0.75 AND petal_width < 1.35 THEN 2
WHEN petal_width >= 0.75 AND petal_width < 1.75 AND petal_width >= 1.35 AND sepal_width < 2.65 THEN 3
WHEN petal_width >= 0.75 AND petal_width < 1.75 AND petal_width >= 1.35 AND sepal_width >= 2.65 THEN 2
WHEN petal_width >= 0.75 AND petal_width >= 1.75 THEN 3
END AS prediction
FROM
iris;
The above SQL will append a prediction column to the selected data, showing the predicted category.
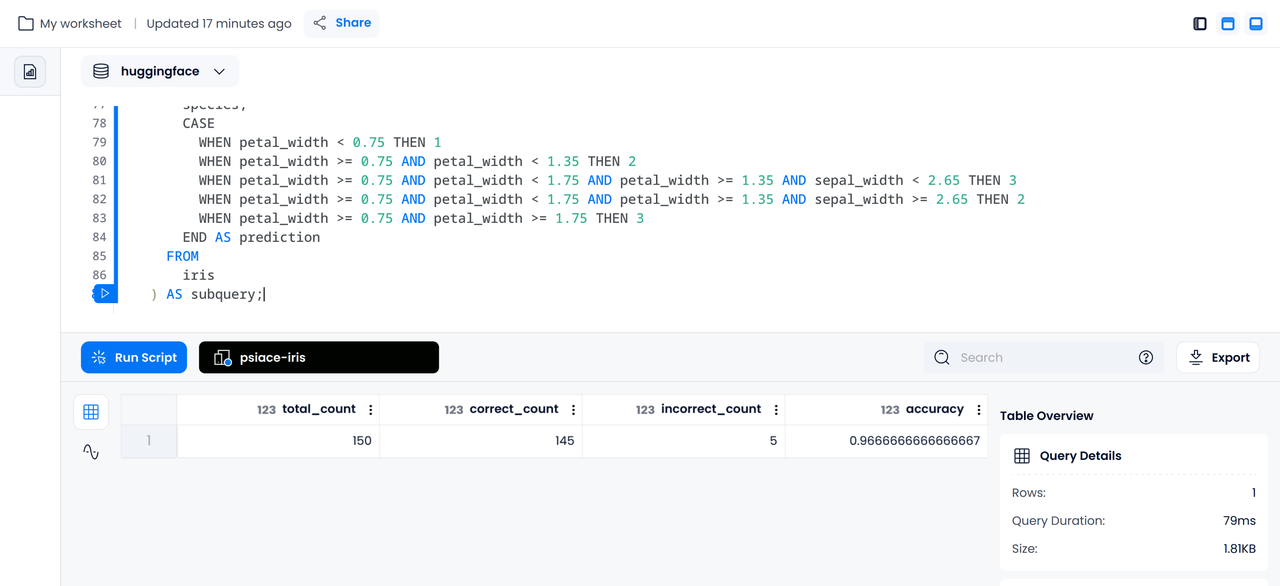
Since the iris dataset already has a category column, we can further calculate the accuracy of the model on the dataset using SQL based on the prediction results:
SELECT
COUNT(*) AS total_count,
SUM(CASE WHEN subquery.species = subquery.prediction THEN 1 ELSE 0 END) AS correct_count,
SUM(CASE WHEN subquery.species <> subquery.prediction THEN 1 ELSE 0 END) AS incorrect_count,
SUM(CASE WHEN subquery.species = subquery.prediction THEN 1 ELSE 0 END) / COUNT(*) AS accuracy
FROM
(
SELECT
sepal_length,
sepal_width,
petal_length,
petal_width,
species,
CASE
WHEN petal_width < 0.75 THEN 1
WHEN petal_width >= 0.75 AND petal_width < 1.35 THEN 2
WHEN petal_width >= 0.75 AND petal_width < 1.75 AND petal_width >= 1.35 AND sepal_width < 2.65 THEN 3
WHEN petal_width >= 0.75 AND petal_width < 1.75 AND petal_width >= 1.35 AND sepal_width >= 2.65 THEN 2
WHEN petal_width >= 0.75 AND petal_width >= 1.75 THEN 3
END AS prediction
FROM
iris
) AS subquery;
Summary
In this article, we demonstrated how to directly access and query Hugging Face datasets using Databend. We also showed how to preprocess data using SQL to conform to data standards. Additionally, by utilizing the
CASE WHEN
All SQL scripts in the post were executed successfully in Databend Cloud. If you're interested in exploring more, just jump in and give it a shot.
Subscribe to our newsletter
Stay informed on feature releases, product roadmap, support, and cloud offerings!



